Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Sync Channel Tutorial#
Note
This is a long-form tutorial on sorting. See here for a quick how-to.
# --- hide: start ---
import shutil
import spikewrap as sw
if (derivatives_path := sw.get_example_data_path("openephys") / "derivatives").is_dir():
shutil.rmtree(derivatives_path)
# --- hide: stop ---
Warning
Currently, the only supported sync channel is from Neuropixels Imec stream (in which it is the 385th channel). Please get in contact to see other cases supported.
Sync channels are used in extracellular electrophysiology to coordinate timestamps from across acquisition devices.
In spikewrap, the sync channel can be inspected and edited. This step
must be performed prior to preprocessing.
Warning
spikewrap does not currently provide any methods for concatenating the sync channel.
This is because a straightforward concatenation may be error prone
(see here for more details).
We are keen to extend sync-channel processing functionality, please get in contact with your use-case
to help usc extend support.
Inspecting the sync channel#
The sync channel data can be obtained as a numpy array, or plot.
Raw data must be loaded prior to working with the sync channel.
The sync channel for a particular run is specified with the run_idx
parameter. Runs are accessed in the order they are loaded (as specified
with run_names, see spikewrap.Session.get_raw_run_names).
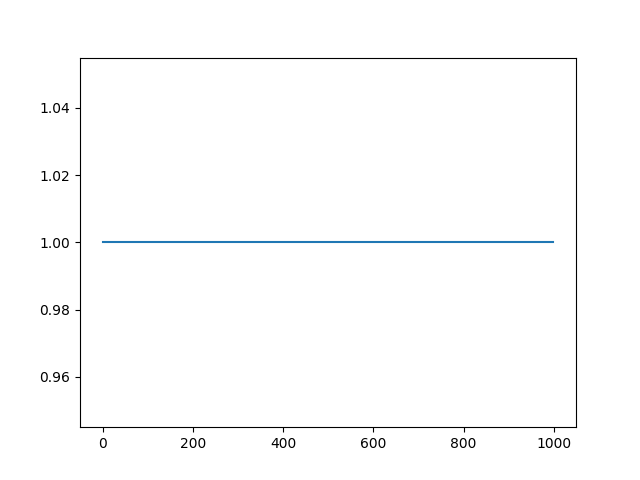
In this toy example data, the sync channel is set to all ones (typically,
it would be all 0 interspersed with 1 indicating triggers).
import spikewrap as sw
session = sw.Session(
subject_path=sw.get_example_data_path("openephys") / "rawdata" / "sub-001",
session_name="ses-001",
file_format="openephys",
run_names="all"
)
session.load_raw_data()
session.get_raw_run_names()
# get the sync channel data as a numpy array
run_1_sync_channel = session.get_sync_channel(run_idx= 0)
# or plot the sync channel
plot = session.plot_sync_channel(run_idx=0, show=True) # TODO: accept the plot and explain. also explain the results
/home/runner/work/spikewrap/spikewrap/spikewrap/process/_loading.py:146: UserWarning: The sessions or runs provided for are not in creation datetime order.
They will be concatenated in the order provided, as:
['recording2', 'recording1'].
warnings.warn(
Loading data from path: /home/runner/work/spikewrap/spikewrap/spikewrap/examples/example_tiny_data/openephys/rawdata/sub-001/ses-001/ephys/Record Node 104/experiment1/recording2
Loading data from path: /home/runner/work/spikewrap/spikewrap/spikewrap/examples/example_tiny_data/openephys/rawdata/sub-001/ses-001/ephys/Record Node 104/experiment1/recording1
Zeroing out the sync channel#
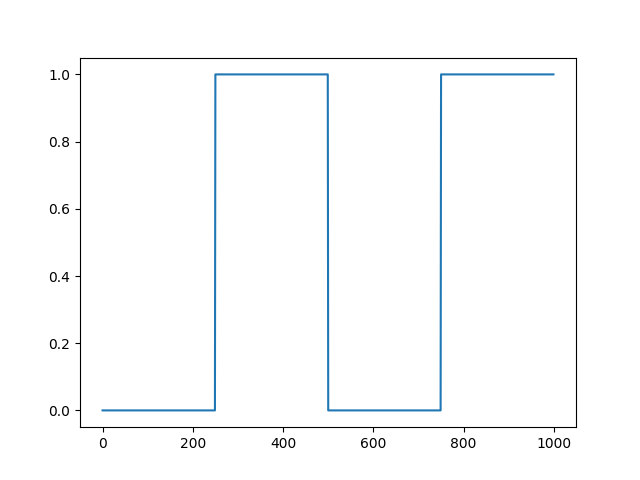
The sync channel can be edited to remove triggers by setting periods of the sync channel to zero:
session.silence_sync_channel(
run_idx=0,
periods_to_silence=[(0, 250), (500, 750)]
)
The function takes a list of 2-tuples, where the entries indicate the start and end of the period to zero (in samples).
Under the hood this uses the spikeinterface function silence_periods(), to zero-out sections of the sync channel.
After plotting the edited sync channel, we see that the periods defined above have been zeroed out:
plot = session.plot_sync_channel(run_idx=0, show=True)
Refreshing the sync channel#
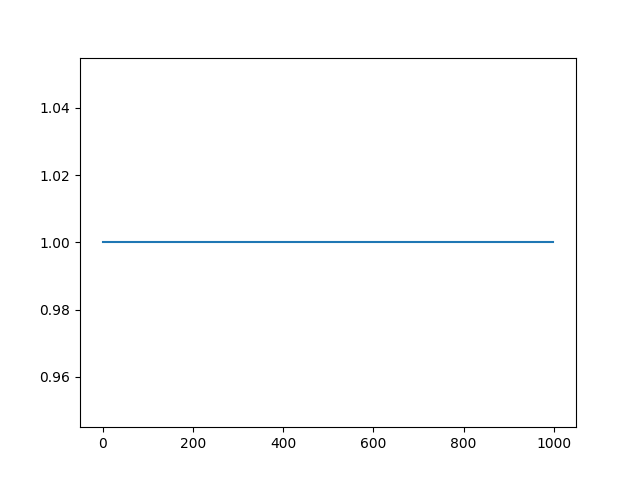
To undo any changes made to the sync-channel, the raw data can be reloaded:
session.load_raw_data(overwrite=True)
# The sync channel is back to original (silenced periods removed)
plot = session.plot_sync_channel(run_idx=0, show=True)
Loading data from path: /home/runner/work/spikewrap/spikewrap/spikewrap/examples/example_tiny_data/openephys/rawdata/sub-001/ses-001/ephys/Record Node 104/experiment1/recording2
Loading data from path: /home/runner/work/spikewrap/spikewrap/spikewrap/examples/example_tiny_data/openephys/rawdata/sub-001/ses-001/ephys/Record Node 104/experiment1/recording1
Saving the sync channel#
The sync channel can be saved with:
session.save_sync_channel(overwrite=False, slurm=False)
Saving sync channel for: recording2...
Saving sync channel for: recording1...
which will save the sync channel for all loaded runs to the run folder. See the SLURM tutorial for more information on using slurm.
Total running time of the script: (0 minutes 0.291 seconds)