Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Overview#
(1 minute read)
spikewrap provides a Python interface for managing and
sharing standardized extracellular electrophysiology pipelines.
Built on SpikeInterface,
spikewrap places emphasis on delivering standardised outputs and
materials for convenient data quality checks.
Attention
spikewrap is currently in a consultation stage, where feedback on the
workflow is being actively solicited. New features—such as sorting,
subject-level analysis, quality checks, support for more probes,
and additional preprocessing steps—are planned for implementation soon.
Please get in contact with feedback and suggestions!
Running spikewrap#
spikewrap expects a project to be organised either in
NeuroBlueprint
format (which is recommended), for example:
└── rawdata/
└── sub-001_.../
└── ses-001_.../
└── ephys/
├── run-001_g0_imec0/
│ ├── run-001_g0_t0.imec0.ap.bin
│ └── run-001_g0_t0.imec0.ap.meta
└── run-002_g0_imec0/
│ ├── ...
└── ...
└── rawdata/
└── sub-001_.../
└── ses-001_.../
└── ephys/
└── Recording Node <ID>/
└── experiment1/
├── recording1/
│ └── ...
└── recording2/
│ └── ...
└── ...
or in custom formats with subject, session and recording folder levels as below:
Supported Custom Organisation
└── root_folder>/
└── my_subject_name/
└── my_session_name/
├── run-001_g0_imec0/
│ ├── run-001_g0_t0.imec0.ap.bin
│ └── run-001_g0_t0.imec0.ap.meta
└── run-002_g0_imec0/
│ ├── ...
└── ...
In the OpenEphys case, the input data would look like:
└── root_folder/
└── my_subject_name/
└── my_session_name/
└── Recording Node <ID>/
└── experiment1/
├── recording1/
│ └── ...
└── recording2/
│ └── ...
└── ...
SpikeGlx or OpenEphys systems with Neuropixels probes are currently supported (see Supported Formats for details).
We can preprocess, visualise and save a recording session with a few function calls:
import spikewrap as sw
subject_path = sw.get_example_data_path() / "rawdata" / "sub-001"
session = sw.Session(
subject_path=subject_path,
session_name="ses-001",
run_names="all",
file_format="spikeglx" # or "openephys"
)
# Run (lazy) preprocessing, for fast plotting
# and prototyping of preprocessing steps
session.preprocess(
configs="neuropixels+kilosort2_5",
per_shank=True,
concat_runs=False,
)
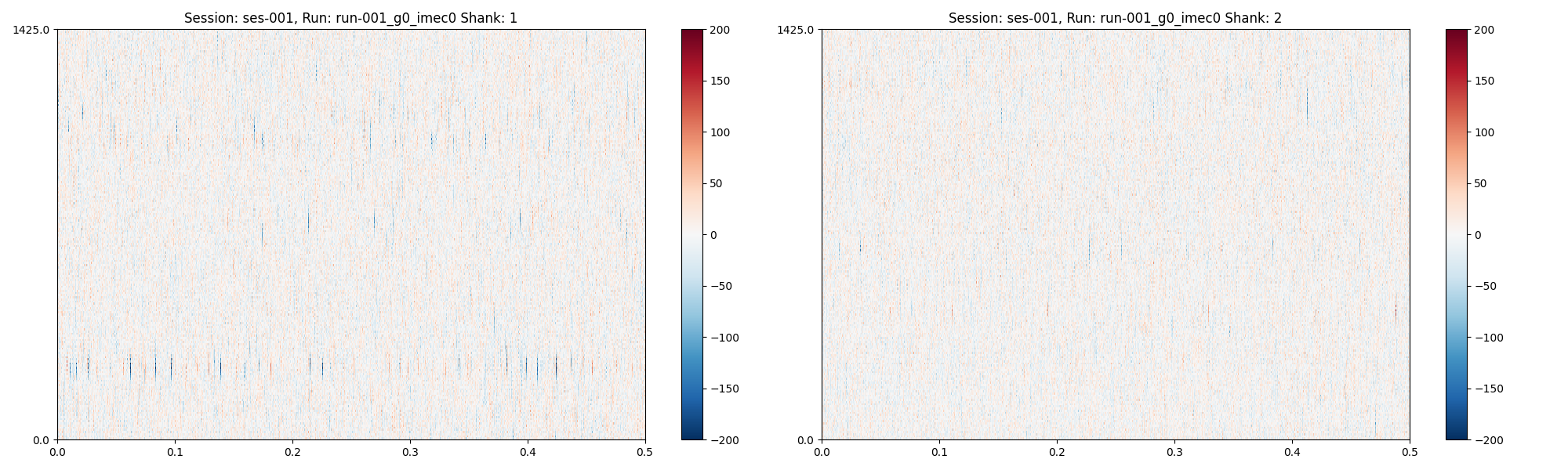
plots = session.plot_preprocessed(
run_idx=0,
time_range=(0, 0.5),
show_channel_ids=False,
show=True
)
# Write preprocessed data to disk, optionally
# in a SLURM job (if on a HPC)
session.save_preprocessed(
overwrite=True,
n_jobs=12,
slurm=False
)
The preprocessing options are: {
"1": [
"bandpass_filter",
{
"freq_max": 6000,
"freq_min": 300
}
],
"2": [
"common_reference",
{
"operator": "median",
"reference": "global"
}
]
}
Loading data from path: /home/runner/work/spikewrap/spikewrap/spikewrap/examples/example_tiny_data/spikeglx/rawdata/sub-001/ses-001/ephys/run-001_g0_imec0
Loading data from path: /home/runner/work/spikewrap/spikewrap/spikewrap/examples/example_tiny_data/spikeglx/rawdata/sub-001/ses-001/ephys/run-002_g0_imec0
Split run: run-001_g0_imec0 by shank. There are 2 shanks.
Split run: run-002_g0_imec0 by shank. There are 2 shanks.
Saving data for: run-001_g0_imec0...
`overwrite=True`, so deleting all files and folders (except for slurm_logs) at the path:
/home/runner/work/spikewrap/spikewrap/spikewrap/examples/example_tiny_data/spikeglx/derivatives/sub-001/ses-001/ephys/run-001_g0_imec0
Saving sync channel for: run-001_g0_imec0...
write_binary_recording
n_jobs=4 - samples_per_chunk=60,000 - chunk_memory=21.97 MiB - total_memory=87.89 MiB - chunk_duration=2.00s
write_binary_recording: 0%| | 0/1 [00:00<?, ?it/s]
write_binary_recording: 100%|██████████| 1/1 [00:00<00:00, 4.68it/s]
write_binary_recording: 100%|██████████| 1/1 [00:00<00:00, 4.67it/s]
write_binary_recording
n_jobs=4 - samples_per_chunk=60,000 - chunk_memory=21.97 MiB - total_memory=87.89 MiB - chunk_duration=2.00s
write_binary_recording: 0%| | 0/1 [00:00<?, ?it/s]
write_binary_recording: 100%|██████████| 1/1 [00:00<00:00, 4.38it/s]
write_binary_recording: 100%|██████████| 1/1 [00:00<00:00, 4.37it/s]
Saving data for: run-002_g0_imec0...
`overwrite=True`, so deleting all files and folders (except for slurm_logs) at the path:
/home/runner/work/spikewrap/spikewrap/spikewrap/examples/example_tiny_data/spikeglx/derivatives/sub-001/ses-001/ephys/run-002_g0_imec0
Saving sync channel for: run-002_g0_imec0...
write_binary_recording
n_jobs=4 - samples_per_chunk=60,000 - chunk_memory=21.97 MiB - total_memory=87.89 MiB - chunk_duration=2.00s
write_binary_recording: 0%| | 0/1 [00:00<?, ?it/s]
write_binary_recording: 100%|██████████| 1/1 [00:00<00:00, 5.47it/s]
write_binary_recording: 100%|██████████| 1/1 [00:00<00:00, 5.47it/s]
write_binary_recording
n_jobs=4 - samples_per_chunk=60,000 - chunk_memory=21.97 MiB - total_memory=87.89 MiB - chunk_duration=2.00s
write_binary_recording: 0%| | 0/1 [00:00<?, ?it/s]
write_binary_recording: 100%|██████████| 1/1 [00:00<00:00, 4.40it/s]
write_binary_recording: 100%|██████████| 1/1 [00:00<00:00, 4.40it/s]
with data output to the standardised NeuroBlueprint structure:
└── root_folder/
└── derivatives/
└── sub-001/
└── ses-001 /
└── ephys/
└── run-001_g0_imec0/
│ ├── preprocessed/
│ │ ├── shank_0/
│ │ │ └── si_recording/
│ │ │ └── <spikeinterface binary>
│ │ └── shank_1/
│ │ └── si_recording/
│ │ └── <spikeinterface binary>
│ └── sync/
│ └── sync_channel.npy
└── run-002_g0_imec0/
└── ...
Next, visit Get Started and Tutorials to try out spikewrap.
Total running time of the script: (0 minutes 1.974 seconds)